[This post was updated on 11-Dec-2024 to reflect the publication of the 'HBDs in drug design' preprint as the K2022 article]
In this post I’ll look at a couple of fragment selection themes with a hydrogen bond donor (HBD) focus. The material has been taken from the recent ‘HBDs in drug design’ preprint (HBD3; this would subsequently be published as the K2022 article) which introduced the term ‘hydrogen bond donor-acceptor asymmetry’ and suggested that we need to think differently about HBDs and hydrogen bond acceptors (HBAs) in drug design. One example of these hydrogen bond donor-acceptor asymmetries is that HBAs are typically more strongly solvated than HBDs in aqueous media and this is especially relevant to lead optimization (as shown in the graphical abstract for HBD3 below).
However, this post is about fragment selection, rather than fixing ADME, and so I’ll say something about differences between HBDs and HBAs in the context of binding to targets. Let’s suppose that you’d like to exploit an HBD in the binding site of your target. All you need to do is place an HBA at a point in space where it can form a good hydrogen bond (taking care to address issues like steric footprint and conformational energy) and you’ve got it sorted. However, life is not quite so simple if you’re trying to exploit an HBA in the binding site because the HBD (e.g., amide NH) that you present to it will almost invariably be accompanied by an HBA (e.g., amide carbonyl O). In contrast, it is relatively easy to design an HBA (e.g., pyridine N) into a ligand structure that is not accompanied by an HBD.
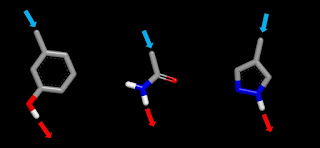
In HBD3, I describe the HBA that accompanies pretty much every neutral HBD as ‘co-occurring’. The problem for designers is that the co-occurring HBA, which is likely to come with a larger desolvation penalty than that for the HBD, needs to be accommodated and this places constraints on design. It’s also more difficult to achieve ‘line-of-sight’ access with HBDs than is the case for HBAs (you’re likely to need line-of-sight access when targeting a polar atom at the bottom of a relatively narrow binding pocket). The following figure should give you a better idea of what I’m getting at and let’s assume that we’re trying to donate an HB to HBA sitting at the bottom of a narrow and otherwise non-polar binding pocket. Although each of the three structures has appropriate geometry for line-of-sight access, things are not likely to end well if you try to exploit this line-of-sight access in a real-life design situation.
Let’s start with the phenol and, although not pertinent to this discussion, it’s worth mentioning that hydroxyl groups are prone to conjugation in phase 2 metabolism (drugs get hydroxylated in phase 1 metabolism in order to facilitate clearance). Donation of an HB by a ligand hydroxyl to a target HBA also brings the hydroxyl oxygen (the co-occurring HBA) into proximity with the molecular surface of the target. This increases the likelihood of an energetic penalty resulting from desolvation of the phenolic oxygen. One subtle point is that donation of an HB by the phenolic hydroxyl increases the HB basicity of the oxygen which effectively increases the energetic cost of desolvating it.
The co-occuring HBA of the primary amide is an even bigger problem than for phenol because the high polarity of the carbonyl oxygen means that it carries a large desolvation penalty (bad news if you’re trying to hit an HBA at the bottom of a narrow and otherwise non-polar binding pocket). If this is not enough of a problem, you also need to worry about desolvation penalties associated with the second HBD (the primary amide has two HBDs and methyl-capping will take out the one that you need for hitting that HBA at the bottom of the binding pocket). As Lady Bracknell might have observed, “One desolvated polar atom may be regarded as a misfortune; to lose solvation of two polar atoms looks like carelessness”.
The last of the trio of structures is pyrazole linked at C4 and this avoids problems that might result from biasing the tautomeric preference. Pyrazole is a great warhead if you’re targeting a proximal HBD and HBA (as is the case when trying to hit a kinase hinge). However, pyrazole’s HBA may become a liability when trying to hit the HBA at the bottom of that otherwise non-polar binding pocket. Why not just take out pyrazole’s HBA, you might ask? The problem is that pyrroles are very electron rich and tend to be quite reactive. One tactic is to move the co-occurring HBA from the ring to the linker (1 and 2) in a way that makes the linker electron-withdrawing and pray for a less destabilizing contact between the co-occurring HBA and the binding site. Alternatively, you can take out the co-occurring HBA and modify the linker to make it more electron-withdrawing (3). I’ve included Hammett σ values in the graphic and these will give you an idea how the substituents vary in their ability to suck electron density out of the pyrrole ring (beneficial both for making the pyrrole ring more rugged and increasing the HB acidity of its NH HBD). I see these fragments as being of about the right size to be screened crystallographically but you might want something a bit larger than methyl if you’re using another detection method.
I’ve made a small selection of substituted guanidines that I think may of interest for screening as fragments. The pKa values that I quote in this post are from an article by two former colleagues (Peter Taylor and Alan Wait who are sadly both deceased) and this is an excellent source of measured guanidine pKa values. Two of these (4 and 5) will be predominantly protonated at neutral pH although there’ll still be a significant amount of the neutral form that you’ll need for permeability. The other two guanidines will be predominantly neutral at neutral pH although 6 is sufficiently basic to protonate in lysosomes. As for the pyrroles, I see these as about the right size to be screened crystallographically but you might want something a bit bigger than methyl if you plan to use a different detection method.